© Borgis - Postępy Nauk Medycznych 4/2015, s. 277-281
*Łukasz Chabros1, 2, Robert T. Kuthan1, 2, Anna Sawicka-Grzelak1, 2, Grażyna Młynarczyk1, 2
Porównanie wyników identyfikacji wankomycyno-opornych enterokoków metodą MALDI-TOF MS, Vitek 2 i API 20 STREP
Comparison of identification methods of vancomycin-resistant enterococci in MALDI-TOF MS, Vitek 2 and API 20 STREP
1Department of Medical Microbiology, The Infant Jesus Teaching Hospital, Warsaw
Head of Department: Anna Sawicka-Grzelak, PhD
2Chair and Department of Medical Microbiology, Medical University of Warsaw
Head of Department: Associate Professor Grażyna Młynarczyk, PhD
Streszczenie
Wstęp. Z klinicznego punktu widzenia, w zakażeniach wywoływanych przez rodzaj Enterococcus, najważniejsze jest różnicowanie gatunków: E. faecium i E. faecalis (odpowiedzialne za ponad 90% wszystkich zakażeń) oraz E. gallinarum, E. casseliflavus i E. flavescens, ze względu na ich naturalną oporność na niskie stężenia wankomycyny (fenotyp VanC). W trakcie badań nad szczepami enterokoków opornych na wankomycynę (VRE) zaobserwowano, że niektóre enterokoki izolowane z materiałów klinicznych od hospitalizowanych chorych były identyfikowane przez systemy oparte na biochemicznej identyfikacji: Vitek 2 i API 20 Strep (bioMereiux, Francja) jako E. gallinarum, podczas gdy inne metody identyfikowały te szczepy jako E. faecium oporne na wankomycynę.
Cel. Celem pracy było porównanie systemu Vitek® 2, API 20 STREP oraz MALDI-TOF MS w identyfikacji gatunkowej enterokoków.
Materiały i metody. 100 szczepów drobnoustrojów z rodzaju Enterococcus, poddano identyfikacji do poziomu gatunku w systemach opartych o rozkład cech biochemicznych (systemy Vitek® 2 i API 20 STREP) oraz o spektrometrię masową (MALDI-TOF). Metodami referencyjnymi były metody genetyczne wykrywające geny ddl i fragmentu genu vanC.
Wyniki. Różnice w identyfikacji enterokoków wynosiły od 2% w systemie VITEK®2 do 6% dla ATBTM Expression względem MALDI – Biotyper oraz metod molekularnych.
Wnioski. Wysoki koszt oraz czasochłonność metoda molekularnych, wyklucza je z zastosowania w rutynowej diagnostyce laboratoryjnej. Natomiast zgodność wyników identyfikacji gatunkowej w spektrometrii masowej MALDI-TOF z metodami genetycznymi, skłania do jej wykorzystania w rutynowej diagnostyce mikrobiologicznej.
Summary
Introduction. From the clinical point of view, differentiation of the following species in the genus Enterococcus is the most important: E. faecium and E. faecalis (responsible for over 90% of all enterococcal infections) and E. gallinarum, E. casseliflavus, E. flavescens due to their resistance to low concentrations of vancomycin (VanC phenotype). During research of enterococci strains resistant to vancomycin (VRE), we are observed some of those strains isolated from clinical specimens from hospitalized patients were identified by systems based on biochemical identification: Vitek® 2 and Api 20 STREP (bioMereiux, France) as E. gallinarum while other methods demonstrated that they were vancomycin resistant E. faecium (VRE).
Aim. Aim of study was to compare system Vitek2 and Api 20 STREP vs MALDI-TOF MS for enterococci identification.
Material and methods. 100 strains of enterococci were identified for species in systems based on biochemical reactions (system Vitek® 2 and API 20 STREP) and mass spectrometry (MALDI-TOF). The detection of ddl and vanC genes was the reference method.
Results. The differences in identification of enterococci ranged from 2% in Vitek® 2 system to 6% for ATBTM Expression system in relation to MALDI – Biotyper.
Conclusions. High cost and time-consuming of molecular methods, exclude them from the application in routine laboratory diagnosis. However consistency of results in species identification in mass spectrometry MALDI-TOF with genetic methods, abet to use its in routine microbiological diagnosis.

INTRODUCTION
Microorganisms of the Enterococcus spp. are Gram--positive, non-spore-forming, relatively anaerobic cocci, in-laying chains or pairs. Most strains could grow at temperatures between 10°C and 45°C, pH 9.6 and in the presence of 6.5% sodium chloride solution (1). They are microorganisms being extremely resistant to harsh environments and are able to survive heating at 60°C for 30 min (1). These bacteria are typical opportunistic pathogens which can cause severe nosocomial infections, they are difficult to treat because of their inherent or acquired resistance to multiple classes of drugs. The term „enterocoque” was for the first time used in the French publication dated 1899 but, due to the morphology of cells, a characteristic image of light microscopy, and the negative reaction to the presence of catalase, to the end of the 70s of the twentieth century enterococci were included among the genus Streptococcus (2-4). In 1984, Schleifer proposed separation of a separate genus Enterococcus spp., basing on the homology analysis of the 16S rRNA sequence between streptococci and enterococci (5). Until now, more than 40 species of the genus Enterococcus have been described, but the following are of clinical relevance: Enterococcus faecium and Enterococcus faecalis, being responsible for over 90% of all infections and Enterococcus gallinarum, Enterococcus casseliflavus and Enterococcus flavescens, due to their natural resistance to low levels of vancomycin (phenotype VanC1, VanC2 and VanC3) (1, 6, 7).
The principle of MALDI-Biotyper operation is based on laser desorption involving matrix (Matrix Assisted Laser Desorption Ionisation – MALDI), consisting of mild ionization using a laser with a specially constructed matrix that absorbs the laser energy to transfer it onto the analyzed protein. Then, a time of flight (Time of Flight – TOF) analyzer – being a mass analyzer – measures the time of flight of ions from the acceleration of the analyzer to the strike at the detector. Ion time of flight is converted to molecular ion ratio of the weight of its electric charge (m/z), and the greater the charge, the longer the ion flight time. The result is obtained as a result of mass spectrum (in the range of 2 to 20 kDa), which gives a unique pattern of protein (molecular fingerprint) compared to a database for identification of organisms (fig. 1). A computer program in the MALDI-Biotyper system, in addition to the mass spectrum also shows the numerical values specifying the degree of identification of the microorganism. Range: 2300-3000 means a very likely, species identification, 2,000-2,299 is a safe identification of the genus and a probable identification of the species, 1,700-1,999 is a likely determination of the genus, and values between 0,000-1,699 represent an incredible identification.
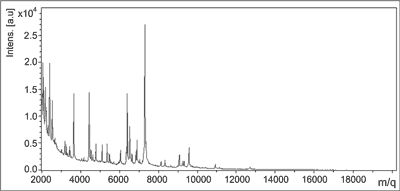
Fig. 1. Sample mass spectrum obtained for E. faecium in the MALDI-Biotyper system (Bruker Daltonics, Germany).
Aim
The aim of the study was to compare three methods for identification of enterococci: biochemical methods (the ATB Expression System and Vitek 2 systems) and mass spectrometry MALDI-TOF.
MATERIAL AND METHODS
One hundred strains of vancomycin-resistant enterococci (VRE) were analyzed, as isolated from patients after kidney or liver problems, staying in one of the teaching hospitals in Warsaw in the years 2010-2011. The strains were grown from rectal swabs (n = 52), urine (n = 17), wound swabs (n = 9), catheters (n = 7), peritoneal fluid (n = 7), bile (n = 4), and blood (n = 3) and a portion of the tissue (n = 1). Pre-selection and identification of clinical primers were performed based on the procedure by Facklam and Collins (8) and included: analysis of the morphology of colonies on Columbia Agar, as supplemented with 5% of defibrinated sheep blood, performing a Gram-stained preparation, test for the presence of catalase and pyrrolidonyl peptidase. Clinical materials were also seeded on D-Coccosel Agar – in order to isolate enterococci decomposing esculin to esculatin in the presence of bile salts. Rectal swabs in the direction of testing the presence of vancomycin – resistant enterococci were also seeded on selective chromID VRE. The bases used were produced by bioMèrieux (France). All of the grown enterococci were subjected to identification by means of the Vitek 2 system and the API 20 STREP biochemical test in the ATB Expression system. All procedures were performed according to the manufacturer’s instructions. For comparison, all the strains were further identified using mass spectrometry (MALDI-TOF) on the MALDI-Biotyper apparatus (Bruker Daltonics, Germany).
Powyżej zamieściliśmy fragment artykułu, do którego możesz uzyskać pełny dostęp.
Mam kod dostępu
- Aby uzyskać płatny dostęp do pełnej treści powyższego artykułu albo wszystkich artykułów (w zależności od wybranej opcji), należy wprowadzić kod.
- Wprowadzając kod, akceptują Państwo treść Regulaminu oraz potwierdzają zapoznanie się z nim.
- Aby kupić kod proszę skorzystać z jednej z poniższych opcji.
Opcja #1
29 zł
Wybieram
- dostęp do tego artykułu
- dostęp na 7 dni
uzyskany kod musi być wprowadzony na stronie artykułu, do którego został wykupiony
Opcja #2
69 zł
Wybieram
- dostęp do tego i pozostałych ponad 7000 artykułów
- dostęp na 30 dni
- najpopularniejsza opcja
Opcja #3
129 zł
Wybieram
- dostęp do tego i pozostałych ponad 7000 artykułów
- dostęp na 90 dni
- oszczędzasz 78 zł
Piśmiennictwo
1. Gillespie S, Hawkey P: Principles and Practice of Clinical Bacteriology. 2 ed., John Wiley&Sons, Ltd, 2006: 60-71.
2. Lancefield RC: A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med 1933; 57(4): 571-595.
3. Schleifer KH, Klipper-Balz R: Molecular and chemotaxonomic approaches to the calssification of streptococcic, enterococcoci and lactococcoci. Syst Appl Microbiol 1987; 10: 1-9.
4. Murray BE: The live and times of the Enterococcus. Clin Microbilogy Rev 1990; 3: 46-65.
5. Schleifer KH, Klipper-Balz R: Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int J Syst Bacteriol 1984; 34: 31-34.
6. Pappas G, Liberopoluos E, Tsianos E, Elisaf M: Enterococcus casseliflavus bacteremia. Case raport and literature review. J Infec Diseases 2004; 48: 206-208.
7. Patterson JE, Sweeney AH, Simms M et al.: An analysis of 110 serious enterococcal infections. Epidemiology, antibiotic, susceptibility and outcome. Medicine (Baltimore) 1995; 74: 191-200.
8. Facklam RR, Collins MD: Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol 1989; 27: 731-734.
9. Clark NC, Teixeira LM, Facklam RR, Tenover FC: Detection and differentiation of vanC-1, vanC-2, and vanC-3 glycopeptide resistance genes in enterococci. J Clin Microbiol 1998; 36(8): 2294-2297.
10. Domig KJ, Mayer HK, Kneifel W: Methods used for the isolation, enumeration, characterisation and identification of Enterococcus spp. 2. Pheno- and genotypic criteria. Int J Food Microbiol 2003; 1; 88(2-3): 165-188.
11. Facklam RR, Carvalho MGS, Teixeira LM: History, taxonomy, biochemical characteristics and antibiotic susceptibility testing of enterococci. [In:] Gilmore MS, Clevell DB, Courvalin PM et al.: The Enterococci: Pathogenesis, Molecular Biology, Antybiotic Resistance. ASM Press, DC: Washington 2002: 1-54.
12. Brigante G, Lozano F, Bettaccini A et al.: Use of the Phoenix automated system for identification of Streptococcus and Enterococcus spp. J Clin Microbiol 2006; 44(9): 3263-3267.
13. d’Azevedo PA, Siquiera I Gumel J et al.: Evaluation of the automated system Vitek 2 for identification and antimicrobial susceptibility testing of Brazilian Gram-positive cocci strains. Braz J Infect Dis 2009; 3(2): 107-110.
14. Eigner U, Schmid A, Wild U et al.: Analysis of the comparative workflow and performance characteristics of the Vitek 2 and Phoenix systems. J Clin Microbiol 2005; 43(8): 3829-3834.
15. McGregor A, Schio F, Beaton S et al.: MicroScan WalkAway diagnostic microbiology system – an evaluation. Pathology 1995; 27(2): 172-176.
16. Sader HS, Biedenbach D, Jones RN: Evaluation of Vitek and API 20S for species identification of enterococci. Diagn Microbiol Infect Dis 1995; 22(4): 315-319.
17. Tritz DM, Iwen PC, Woods GL: Evaluation of MicroScan for identification of Enterococcus species. J Clin Microbiol 1990; 28(6): 1477-1478.
18. Koneman EW, Procop GM, Washington W et al.: Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. 6 ed., Lippincot Williams&Wilkins 2006: 700-745.
19. Fang H, Ohlsson AK, Ullberg M, Ozenci V: Evaluation of species-specific PCR, Bruker MS, VITEK MS and the VITEK 2 system for the identification of clinical Enterococcus isolates. Eur J Clin Microbiol Infect Dis 2012; 31(11): 3073-3077.
20. Griffin PM, Price GR, Schooneveldt JM et al.: Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J Clin Microbiol 2012; 50(9): 2918-2931.
21. Crider SR, Colby SD: Susceptibility of enterococci to trimethoprim and trimethoprim-sulfamethoxazole. Antimicrob Agents Chemother 1985; 27(1): 71-75.
22. www.eucast.org.

