© Borgis - New Medicine 4/2012, s. 116-121
*Paweł Kowalczyk1, Karol Chalimoniuk2, Agnieszka Danielak2, Dorota Dziedziela2, Paulina Jankowska2, Marzena Kowalska2, Joanna Laskowska2, Marzena Rachocka2, Jarosław Szczepaniak2, Tomasz Walter2, Paulina Strzyga2, Justyna Szymańska2, Mariusz Słomka2, Katarzyna Zawadka2, Martyna Staszewska2
M13mp18 phage model as a tools of research mutagenic and cytotoxic biological and environmental compounds
1Autonomous Department of Microbial Biology, Faculty of Agriculture and Biology, Warsaw University of Life Sciences, Poland
Head of Department: assoc. prof. Małgorzata Łobocka, MD, PhD
2Scientific Circle of Molecular Biology, Autonomous Department of Microbial Biology, Faculty of Agriculture and Biology, Warsaw University of Life Sciences
Summary
Introduction. Vectors of the mp series are all derived from a recombinant M13 bacteriophage (M13mp1) that carries a short segment of E. coli DNA in its major intergenic region. This segment contains the regulatory sequences and the coding information for the first 146 amino acids of the β-galactosidase gene (lacZ). M13 is an ideal vector for obtaining single-stranded templates required for the dideoxynucleotide chain termination method of sequencing DNA and site-directed mutagenesis using synthetic oligonucleotides.
Aim. M13 phage is a usefull tools for detection of different chemical environmental compounds such as: vinyl chloride (CV), chloroacetaldehyde (CAA) and other alkylating and oxidative agents e.g. trans-4-hydroxy-2-nonenal (HNE).
Material and methods. This vector allow estimate how strong is the chemical compound; by measure the level of survival and mutation frequency in the after transfection into bacterial strains after modification of phage DNA by different compounds concentration.
Results. Modification of single stranded M13 phage DNA with HNE may inhibit in vitro DNA synthesis by T7 DNA polymerase. The most reactive base within the M13 lacZ gene was 63% of guanine residues within the examined sequence), followed by cytosine (55%), adenine (39%), and finally thymine (25%).
Conclusions. The long chain HNE adducts to DNA bases arrest DNA synthesis to all four DNA bases and cause recombination, base substitutions and frameshift mutations in ssDNA and may constitute a strong hindrance for DNA synthesis. This DNA modification may lead to mutation and finally induce tumor progression.
INTRODUCTION
Lac Z Operon
The lac operon is composed of two distinct regions of DNA. These are typically known as the LacI region and the LacZ regions. The LacI region is responsible for controlling the production of ß-galactosidase, the key enzyme that can break down lactose into glucose and galactose. As the LacI region is a gene, it codes for a specialized protein known as a repressor. The LacZ region consists of three seperate gene sequences: lacA, lacY and lacZ (Fig. 1). The lacA gene codes for transacetylase, which seemingly has no role in this system. The lacY gene codes for a lactose permease, which facilitates the movement of lactose into the cell membrane from the outside environment. The lacZ gene codes the ß-galactosidase. This enzyme hydrolyzes the bond between the two sugars, glucose and galactose. If lactose is not available in the medium, the genes for metabolizing the sugar are not expressed. However, if lactose is present in the medium, the genes for metabolizing this sugar are expressed and the bacterium is able to use lactose as an energy source. Preceding each gene region is a control sequence of DNA known as a promoter. In addition to this promoter, the LacZ region also contains an additional region known as an operator (fig. 1).
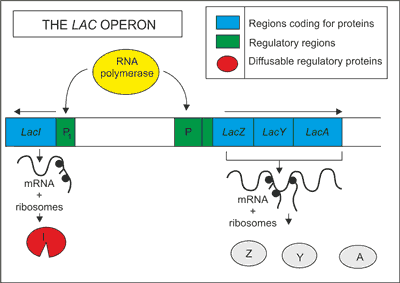
Fig. 1. Schematic presentations of the lac operon.
M13 as a cloning vector and a model for studies of mutations
The lacZ operon has been incorporated into several cloning vectors, specific to E. coli including filamentous bacteriophage e.g. M13. Is a filamentous, male-specific coliphage composed of a single-stranded closed circular molecule of DNA approximately 6400 nucleotides in length and a protein coat. Because of its filamentous structure, additional DNA can be inserted into its genome and packaged into an extended phage capsid. During infection, the phage attach only to the sex pili (conjugation tube) encoded by the F episome and only male bacteria are used to propagate the virus. Bacterial strains carrying F’ episomes and a number of genetic markers useful in work with M13 vectors have been constructed by Messing (1). The most important of these markers are: lacZ?M15A deletion mutant lacking the sequences of the lacZ gene coding for the amino-terminal portion of β-galactosidase (2). The peptide expressed by ?M15 can take part in α-complementation JM105 E. coli strain, a host for bacteriophage M13 vectors carries this deleted version of the lacZ gene on an F’ episome. In phage particles the chromosome of phage exists as a single-stranded circular DNA and is converted by cellular enzymes into a double-stranded circular form, called replicative form (RF). The replicative form replicates and gives rise to single stranded (the + strand) molecules which are packaged into phage and extruded from the cell. M13 does not lyse its host; rather, the phage particles containing single-stranded DNA are continuously secreted into the media by the cells.
Vectors of the mp series are all derived from a recombinant M13 bacteriophage (M13mp1) that carries a short segment of E. coli DNA in its major intergenic region. This segment contains the regulatory sequences and the coding information for the first 146 amino acids of the β-galactosidase gene (lacZ). Messing and others (1) further modified the phage M13mp1 by creating an EcoRI restriction site within the gene encoding β-galactosidase and added different other a useful series of densely packed restriction sites into the lacZ, and created the M13 vectors known as M13mp18. This phage (M13mp18) is easily screened for the introduction of a foreign DNA fragment by assaying for β-galactosidase activity in appropriate E. coli hosts. The properties described above make M13 an ideal vector for obtaining single-stranded templates required for the dideoxynucleotide chain termination method of sequencing DNA and site-directed mutagenesis using synthetic oligonucleotides. The M13mp18 genes are shown (fig. 2) on the map (M13 genes are transcribed clockwise).
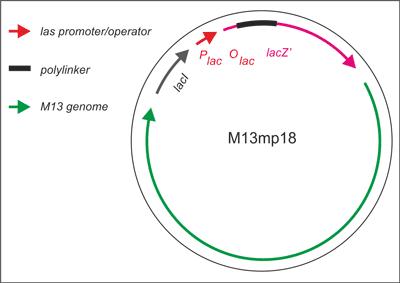
Fig. 2. Development of M13 into a cloning vector.
M13 was developed into a useful cloning vector by inserting the following elements into the genome: a gene for the lac repressor (lacI) protein to allow regulation of the lac promoter: the operator-proximal region of the lacZ gene (to allow for α-complementation in a host with operator-proximal deletion of the lacZ gene): a lac promoter upstream of the lacZ gene; a polylinker (multiple cloning site) region inserted several codons into the lacZ gene.
This fragment, whose synthesis can be induced by IPTG, is capable of intra-allelic (α-complementation with a defective form of beta-galactosidase encoded by host (mutation lacZ?M15). The defective β-galactosidase protein produced by the F episome can complement the defective β-galactosidase encoded on the M13 vector (even with the presence of the polylinker). Ligation of DNA into the polylinker insertionally inactivates the vector-based defective lacZ gene product, disrupting the “α-complementation” leading to the loss of β-galactosidase activity. Thus the plaques retain their normal white color on X-gal/IPTG indicator plates. Screening for “white” plaques amongst the “blue” ones allows selection for DNA molecules inserted into the vector. However, one should be aware that almost any change in the polylinker (such as a disruption of one of the restriction sites) will also give “white” plaques, as will cells which have lost the F’ episome (when selection is relaxed). IPTG in the lacZ gene induces production of the functional galactosidase which cleaves X-Gal and results in a blue colored metobolite. The usual substrate for the lacZ gene protein product is lactose, which is metabolized into galactose and glucose. X-Gal is a colorless, modified galactose sugar, which, when hydrolysed by β-galactosidase yields blue phage plaques. M13 phage system is also an ideal forward mutation system, which can detect base substitutions and frameshits within M13 lacZ gene fragment, that effect β-galactosidase activity and in consequence α-complementation. Mutant phages defective in β-galactosidase can be thus easily identified as white or light blue plaques among wild type dark blue ones. The system enables also to monitor recombination events between lacZ gene positioned in the phage DNA and the host F’ factor, which appear as double deletion of large lacZ gene fragments – a 93 nucleotides correspoding to ?M15, and a 54 nucleotides, which corresponds to polylinker, cloned into M13 vectors.
AIM
The aim of study was to provide a broad database on formation, repair and genotoxic properties of exocyclic DNA adducts. These types of DNA lesions may be induced both by lipid peroxidation (LPO). LPO produces a family of propano-type and etheno-type DNA adducts usually substituted with alkyl chain of different length. The identity of DNA lesions, as well as proteins engaged in their processing are not well elucidated. I intended to investigate the consequences and sequence specificity of interaction with DNA of two types of compounds introducing into DNA exocyclic DNA adducts: (i) one of the major LPO product, trans-4-hydroxy-2-nonenal (HNE). Although HNE is abundant in tissues and plasma of rodents and humans, the molecular characterization of its adducts to DNA bases is still limited. This prompted me to address questions concerning: (I) the identity of HNE adducts to DNA bases by the effect on DNA synthesis in vitro and in transformation into E. coli cells; (II) their mutagenic properties in M13 phage system in lacZ gene.
MATERIAL
Acrylamide, bisacrylamide, calcium chloride, chloroform, PEG, phenol, set of four individual dNTPs and Tris were purchased from Sigma. DNA sequencing kit (T7 Sequencing Kit), [α-S35]ATP (1000 Ci/mmol) and [γ-32P]ATP (3000 Ci/mmol) were from Amersham-Pharmacia Biotech or ICN. IPTG was from Promega. X-gal were obtained from MP Biomedicals.
The compound trans-4-hydroxy-2-nonenal (HNE) was synthesized in the form of a dimethylacetal derivative according to Chandra and Srivastava (6) with minor modifications with prof. Kuśmierek group of IBB PAS. The enzymes: T7 polymerase (Sequenase version 2.0) was obtained from Amersham, Tag polymerase was obtained from Promega.
METHODS
Purification of M13mp18 phage DNA
Bacteriophage M13mp18 was grown overnight at 37°C in JM 105 strain of E. coli in 2YT medium (3). Phage particles were precipitated from the medium with polyethylene glycol, and DNA was isolated by the phenol/ /chloroform method as described by Messing (1).
Modification of M13mp18 phage DNA by e.g. HNE
Single-stranded M13 phage DNA was incubated with different concentrations of HNE at pH 5.5 for 2 h at 37°C. For each HNE concentration, as well as for the untreated control, 20 μg of phage DNA in a total volume of 100 μl was used. Subsequently, the DNA was ethanol precipitated and resuspended in 100 μl of sterile water.
Preparation of competent cells and transformation
Bacteria were grown at 37°C in LB medium and made competent by the CaCl2 method (3). Transfection was performed according to Sambrook (3) with 100 ng phage DNA used to transfect 100 μl of competent cells. Transfection mixtures were plated on LB solid medium with 3 ml of LB soft agar suplemented with 0.4 mM IPTG and 0.5 mg/ml X-gal. Plates were incubated overnight at 37°C and plaques of phage infective centers were scored.
Collection and sequencing of lacZ mutants
M13 lacZ mutants lacking α-complementation and exhibiting low or no β-galactosidase activity were identified as colorless or light blue plaques on LB plates containing IPTG and X-gal. Mutant DNA was isolated acording to Messing (1) and the M13mp18 lacZ gene was sequenced using the dideoxynucleotide chain termination method (4). A T7 Sequencing Kit, [α35 S]-dATP and a specific 19 nucleotide primer, annealing at base positions for amino acids 76-70 of the lacZ gene of the M13 + strand, were used for DNA sequencing. To ensure that mutants were not derived from clonal expansion, eight independent HNE modifications of the phage DNA were performed. Only one transformation was carried out after each DNA modification, and transfection mixtures were plated onto 4 plates. Mutants were collected only from plates treated with 2 mM HNE. Usually only one mutant (three at maximum) was picked up from each plate.
In vitro primer extension on HNE-pretreated M13mp18 phage DNA template
Single stranded M13 phage DNA incubated with HNE (pH 5.5, 2 h, 37°C) was annealed with the 19 nucleotide primer. The primer was labeled for 5 min at 17-20°C by addition of an excess (1-2 μl) of [α35S]-dATP5’ (1000 Ci/mmol) in a mixture for sequencing reactions containing T7 DNA polymerase (4 units per sample). Subsequently, four dNTPs, each at a final concentration of 80 μM, were added and the samples were incubated for 10 min at 37°C for chain elongation. The reaction was terminated by addition of formamide dye, and the products were analyzed by electrophoresis on a sequencing gel. Unmodified phage DNA used as a reference ladder was sequenced by the Sanger method (4) with a T7 Sequencing Kit.
RESULTS
Effect of HNE-DNA adducts on DNA synthesis in vitro
Modification of single stranded M13 phage DNA with HNE (2 mM, 2 h, 37°C) results in formation of template adducts, which inhibit in vitro DNA synthesis by T7 DNA polymerase (fig. 3). The formation of these adducts appeared to be sequence-specific, although no sequence-specificity rule could be proposed. Judging from the frequency of premature chain terminations (the number of DNA polymerase “pause sites”), the most reactive base within the M13 lacZ gene was guanine (pause sites were observed at 63% of guanine residues within the examined sequence), followed by cytosine (55%), adenine (39%), and finally thymine (25%). These results show that adducts to all four DNA bases constitute a strong hindrance for DNA synthesis.
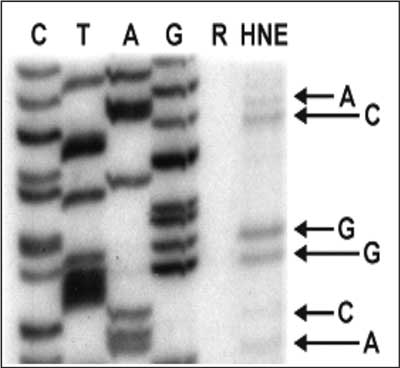
Fig. 3. Primer extension by T7 DNA polymerase in the presence of all four dNTPs on a M13 phage template modified with HNE. Lanes C, A and G correspond to a reference sequence. Lane R shows the products of DNA synthesis performed on a non-modified template. Lane HNE shows the products of DNA synthesis on M13 DNA modified with HNE (2 mM HNE, 2 h, 37°C, pH 5.5). Arrows point to premature chain terminations opposite G, C, A and T sites in the template.
Mutagenic properties and specificity of HNE in M13mp18 phage model
Modification of phage DNA with HNE resulted in E. coli strain decrease of phage survival in the wild type JM105 strain in a dose dependent manner an up to 20-fold and increase of mutation frequency in the M13 lacZ gene (fig. 4. A, B).
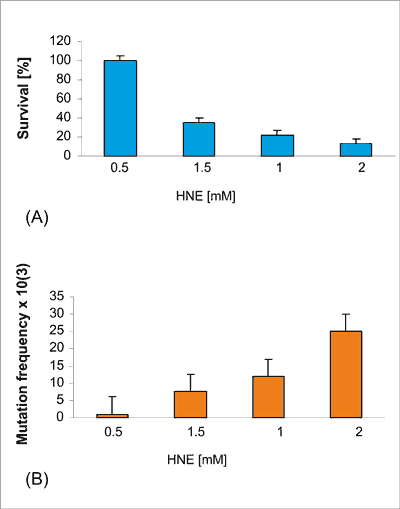
Fig. 4. Survival of M13 phage (Panel A) and mutation frequency (Panel B) in the lacZ gene after modification of phage DNA with HNE (0.1-2 mM) for 2 h at 37°C at pH 5.5 and transfection into E. coli JM 105 strain.
Modification of phage DNA with HNE resulted in a decrease of phage survival in the wild type JM105 strain and an increase in a dose dependent manner, of mutation frequency in the M13 lacZ gene (fig. 4 A, B). The M13 lacZ mutants were collected and sequenced.
The most numerous events found were recombinations between lacZ gene sequences in M13 and the E. coli F’factor DNA. The recombinations were detected as simultaneous deletions of 93 (?M15 deletion of F’lacZ) and 54 (M13 polylinker) nucleotide fragments of phage DNA (tab. 1). These mutants were additionally bearing an A→G transition in the EcoRI site, a base change representing the original base altered in the construction of M13mp phages, which strongly suggests their recombination origin. Thefrequency of these events in HNE-treated phage increased 100-fold in comparison to the rate of spontaneous recombinations. Base substitutions and frameshifts occurred at approximately similar percentage. The major base substitution was the C→T transition, present in 18% of all mutations. Although the C→T transition is also a predominant spontaneous mutation, its frequency in HNE-treated phage increased by almost an order of magnitude (8). Twenty-two percent of point mutations was in the form of frameshifts (8). The frequency of HNE-induced frameshift mutations was about 30-fold higher than that in the spectrum of spontaneous mutations. Among the frameshifts, additions of a single C or G, as well as deletions of a single A or G were observed (tab. 1).
Table 1. Types and frequencies of mutations in the lacZ gene of M13mp18 phage induced by HNE in JM105 strain in comparison with those arising spontaneously in E. coli JM105 strain.
| Type of mutation | HNE induced-wt | Spontaneous |
| Base substitution | Number of mutants | Mutationfrequency
x10-5 | % | Number of mutants | Mutationfrequency
x10-5 | % |
| A→G | 0 | 0 | 34 | 4 | 7.0 | 64 |
| A→C | 5 | 49 | 1 | 1.8 |
| G→C | 6 | 59 | 2 | 3.5 |
| G→T | 1 | 10 | 3 | 5.3 |
| G→A | 0 | 0 | 0 | 0 |
| C→T | 16 | 158 | 11 | 19.3 |
| T→C | 2 | 20 | 0 | 0 |
Frameshifts
insertions: | | | | |
| +G | 4 | 39 | 22 | 0 | 0 | 9 |
| +C | 5 | 49 | 0 | 0 |
| +T | 1 | 10 | 0 | 0 |
| Deletions: | | | | |
| -A | 6 | 59 | 1 | 1.8 |
| -G | 3 | 30 | 1 | 1.8 |
| -T | 0 | 0 | 1 | 1.8 | |
| Recombination deletion of 93+54 nucleotides: | 38 | 374 | 44 | 2 | 3.5 | 6 |
| Other deletions: | 0 | 0 | 0 | 7 | 12.3 | 21 |
| Total: | 87 | 1040 | 100 | 33 | 58 | 100 |
In ssDNA pretreated with HNE, DNA synthesis by T7 DNA polymerase was stopped in a sequence-dependent manner at G ≥ C > A and T sites. HNE increased the mutation rates in the lacZ gene of M13 phage transfected into wild type Escherichia coli (8). The most frequent event was the recombination between lacZ gene sequences in M13 and the E. coli F’ factor DNA. Base substitutions and frameshifts were also observed in approximately similar numbers. Over 50% of base substitutions were the C→T transitions, followed by the G→C and AsyC transversions. In ssDNA the predominant form of HNE-dG adducts is a closed form (8). This reversible reaction can affect the rate of mutations as judged from the studies on related lesions, γ- and α-hydroxy-1,N2-propano--2’-deoxyguanosine (7, 8). Feng and coworkers (7) observed also a large fraction of tandem mutations, suggesting that they are derived from intrastrand crosslinks. No tandem mutations have been observed in my study. Formation of cross-links is very unlikely in ss DNA incubated with HNE for a short period (2 h in comparison with 30 h in 37°C) and immediately transfected into bacteria.
DISCUSSION
Here is shown that HNE adducts to all four DNA bases inhibit DNA synthesis and induce point mutations as well as recombination in ssDNA of the M13 phage. HNE reacts readily with cellular thiols, proteins and nucleic acids, being the most cytotoxic and least mutagenic among the aldehydes produced by LPO. In mammalian cells, only a two to five-fold increase in mutation frequencies could be attained after HNE treatment due to high toxicity that enabled only low doses (maximum 45 μM) to be applied. To avoid the effect of cytotoxicity and to establish the mutagenic potency of HNE adducts to defined DNA bases, we used M13 phage system, in which DNA was modified in vitro and transfected into E. coli cells. This enabled me to apply higher HNE concentrations (up to 2 mM) resulting in an up to 20-fold increase in total mutation frequencies. It has been shown in the literature data that guanine reacted most readily with HNE, followed by C>A>>T. Thymidine appeared to be less sensitive to HNE modification than other bases and accordingly less mutations were targeted to thymine residues than to other bases in the M13 phage.
However, most mutations in wild type E. coli were found at cytosine sites in the template, and these were predominantly the C→T transitions and single cytosine residue additions. Frequent frameshift mutations, mostly C additions within C runs were also observed in M13MB102 phage modified with malondialdehyde (MDA) (5). MDA produces a 3-carbon atom linear adduct to exocyclic nitrogen atom in cytosine. Since, HNE-dC adducts also bear six- or seven-carbon atom side chains, they could exert a similar effect on the active center of the DNA polymerase as the MDA adducts.
CONCLUSIONS
We conclude that long chain HNE adducts to DNA bases arrest DNA synthesis to all four DNA bases and cause recombination, base substitutions and frameshift mutations in ssDNA which are localized mainly at cytosine sites in M13 phage lacZ gene, followed by G and A sites. HNE-DNA adducts are probably engaged in carcinogenesis process via damaging critical genes at mutation hot-spots possibly contributing to genome instability.
Piśmiennictwo
1. Messing J: New M13 vectors for cloning. Methods Enzymology 1983; 101: 20-78. 2. Beckwith JR: A deletion analysis of the lac operator region in Escherichia coli. J Mol Biol 1964; 78: 427-430. 3. Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Labolatory Manual, Cold Spring Harbor Labolatory Press 1989; Cold Spring Harbor. 4. Sanger F, Coulson AR, Barell BJ et al.: Cloning in single stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol 1980; 143: 161-178. 5. Chaudhary M, Benamira K, Johnson A et al.: Induction of mutations by replication of malondialdehyde-modified M13 DNA in Escherichia coli: determination of the extent of DNA modification genetic requirements for mutagenesis, and types of mutations induced. Carcinogenesis 1995; 16: 93-99. 6. Chandra A and Srivastava SK: A synthesis of 4-hydroxy-2- -nonenal and 4-(3H) 4-hydroxy-2-trans-nonenal. Lipids 1997; 32: 779-782. 7. Feng ZH, Hu WW, Amin S et al.: Mutational spectrum and genotoxicity of the major lipid peroxidation product trans-4-hydroxy-2-nonenal induced DNA adducts in nucleotide excision repair-proficient and -deficient human cells. Biochemistry 2003; 42: 7848-7854. 8. Kowalczyk P: The influence of exocyclic DNA adducts In bacterial and mammalian genome instability. New Medicine 2012; 3: 68-73.



