*Marcin Ciszewski1, Tomasz Czekaj1, Piotr Chojnacki2, Eligia M. Szewczyk1
New bacterial zoonotic pathogens
1Department of Pharmaceutical Microbiology and Microbiological Diagnostics, Medical University of Łódź, Poland
Head of Department: prof. Eligia M. Szewczyk, PhD
2Chair of Labour and Social Policy, Faculty of Economics and Sociology, University of Łódź, Poland
Head of Department: prof. Bogusława Urbaniak, PhD
Summary
The article presents the problem of new zoonotic bacterial pathogens posing a threat to humans. Currently, 11 pathogens responsible for causing human zoonotic diseases are being monitored by the European Union epidemiological authorities, 7 of which are bacteria: Salmonella spp., Campylobacter spp., Listeria monocytogenes, Mycobacterium bovis, Brucella spp., Escherichia coli VTEC/STEC and Coxiella burnetti. Nonetheless, many new emerging zoonotic bacteria, which are not currently monitored by ECDC might also pose a serious epidemiological problem in the foreseeable future: Streptococcus iniae, S. suis, S. dysgalactiae subsp. equisimilis and staphylococci: Staphylococcus intermedius and S. pseudintermedius. These species have just crossed the animal-human interspecies barrier. The mechanism of this phenomenon remains unknown. It is connected, however, with genetic variability and capability to survive in changing environment, which are the result of DNA rearrangement and horizontal gene transfer between bacterial cells. The recent substantial increase in the number of scientific publications on this subject testifies to the importance of the problem.
There are approximately 1400 pathogens which currently pose a threat to humans. Most of these pathogens are of zoonotic origin. Among them, there are the most dangerous pathogens, which broke the animal-human interspecies barrier in recent times. E.g. viruses like: HIV, H5N1 and H1H1 influenza, coronaviruses or Ebola have become a serious epidemiological problem during the last 50 years. The number of pathogenic viruses increases by approximately 25 every 10 years. Among bacteria, Helicobacter pylori, pathogenic serotypes of Escherichia coli (VTEC, EIEC, EPEC, EAggEC, DAEC, O104:H4) and Borrelia burgdorferi have emerged during last 50 years.
The primary reservoir of many species is the environment (e.g. Legionella pneumophila, Bacillus anthracis) but most of them (nearly 1000 species and varieties) originate from animal pathogens. Only about 3.5% of all etiological factors are exclusive to humans and nowadays do not have any animal or environmental reservoir. This group comprises e.g. Streptococcus pyogenes, Neisseria gonorrhoeae and Treponema pallidum (1). The notion „zoonoses” is usually used to describe infections that can be transmitted directly from animals to humans. Sometimes, when a pathogen gains the ability of human-to-human transmission, we forget about its animal origin. However, it is still able to infect its primary hosts and might be transported by them. Some zoonotic bacteria have over the years achieved the ability to cause epidemic diseases, like plague (caused by Yersinia pestis) and typhoid fever (caused by Rickettsia prowazekii). New zoonotic human RNA viruses are the most frequently noted, due to their spectacular epidemics, e.g. HIV (transmitted from apes), SARS (from civets), H5N1 influenza (from birds), H1N1 influenza (from pigs) and, last but not least, the most current – Ebola virus (from bats). According to Woolhouse and Gaunt (2), the danger of new zoonotic pathogens emergence (or new varieties of already known pathogens) is one of the biggest challenges of XXI century. Taylor et al. (1) proved that animal pathogens (bacteria and fungi) pose a three-fold higher danger of becoming new etiological factor of human diseases than bacteria and fungi of other origin.
The process of evolutionary changes in the transformation of an animal pathogen into a specialized pathogen of humans has already been described by Wolfe et al. (3). Five steps constituting this process are shown in figure 1. It also demonstrates the current location of the emerging pathogens described in the article. The bases of this evolutionary process usually remain unknown but it is connected with genetic variability and obtaining new features which allow bacteria to survive and spread in a new environment. This ability mainly results from horizontal gene transfer (HGT) and DNA rearrangements (4).
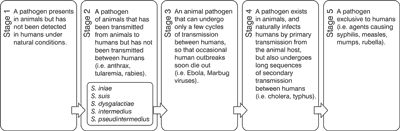
Fig. 1. Scheme presenting evolutionary transformation of zoonotic pathogens, including emerging zoonotic bacterial pathogens described in this article (3 – modified).
European Union epidemiological authorities collect data on zoonoses from all Member States. Their annual reports describe 11 most important zoonotic etiological factors, 7 of which are bacterial pathogens. European Union summary reports on zoonoses include following bacterial pathogens: Salmonella spp., Campylobacter spp., Escherichia coli O104:H4 (VTEC/STEC), Listeria monocytogenes, Mycobacterium bovis, Brucella spp. and Coxiella burnetti (5). These bacteria are well-known and described. However, the abovementioned list includes only some of zoonotic pathogens, selected arbitrarily by the epidemiological authorities, mainly connected with foodborne diseases. Over the last years many other animal pathogens have gained the ability to infect humans. Highlighting the presence of these new bacterial pathogens, as well as the fact that the EU list of monitored pathogens is no longer up-to-date, seems to be of vital importance.
A prevalent genus actively adapting to human environment is Streptococcus. Streptococcus iniae used to be regarded as a significant fish and dolphin pathogen. In the late nineties human infections were also noticed with a large variety of severe symptoms like cellulitis (6), endocarditis (7), meningitis and sometimes sepsis with streptococcal toxic shock syndrome (7). Recent data show that during the past few years over 2000 human S. iniae infections have been noted in Australia and Canada (8, 9). Other Streptococcus species – for instance S. suis, was regarded as an important swine pathogen, connected with high mortality infections. In 2005 the first outbreak of epidemic in humans took place in China. According to the official data 39 people died. Since then, hundreds of cases have been documented, also in many countries in Europe, including Poland. 72.5% of infections led to meningitis, 73.0% of which ended in deafness. 24.2% of patients suffered from sepsis with streptococcal toxic shock syndrome. These statistics show that the frequency of severe infections was significantly higher than the one connected with well-described human streptococci (4, 10, 11). However, the infections of both S. iniae and S. suis are still regarded as primary infections, which cannot transmit between humans (stage 2 according to Wolfe et al.) (9). Usually the route of infection is a direct contact with an infected animal or its meat when a pathogen penetrates the human organism through wounds (12).
Not only pigs or fish might be the reservoir of zoonotic pathogens. Also pets, such as cats and dogs frequently transmit various streptococci and staphylococci. For instance, until recently Streptococcus dysgalactiae had been reported as a typical animal pathogen, not found in clinical samples from humans. In the past few years, more and more frequently, S. dysgalactiae subsp. equisimilis has been isolated from purulent infections in humans, like pharyngitis, pneumonia and necrotizing fasciitis. S. dysgalactiae subsp. equisimilis has also appeared to have the potential to cause focal infections such as sepsis, endocarditis or meningitis (13, 14). Bacteria are usually transmitted by direct contact with an animal or its secretions. Similarly to S. iniae and S. suis, the human-to-human transmission has not been proven yet.
Staphylococcus intermedius frequently colonizes skin and mucosal membranes of dogs and might cause deep pyoderma. The first case of human infection as a result of a dog bite was reported in 1989 (15). Furthermore, Guardabassi et al. (16) demonstrated that multi-resistant S. intermedius strains, infective for dogs, were simultaneously isolated from the oral and nasal cavities of their owners. Human infections are usually mild. However, in patients with compromised immunity they might lead to endocarditis (17), pneumonia (18), brain abscess (19) and acute meningitis (20). The human-to-human transmission of S. intermedius has not been observed yet.
Staphylococcus pseudintermedius also colonizes animal skin and mucosal membranes and might cause opportunistic diseases, like superficial and deep pyoderma or otitis media (21). According to Kadlec et al. (22). Staphylococcus pseudintermedius might also cause septicemia, nephritis, rhinitis and wound infections in cats. The animal-to-human transmission usually results from an animal bite or close contact with a cat or a dog. Similarly to S. intermedius, the colonization of dog owners’ nasal cavity by S. pseudintermedius strains isolated also from their dogs, has been proven (23). This pathogen may cause mild opportunistic infections in humans. However, single cases of sinusitis (24), endocarditis (25) and pneumonia (26) have also been described. As the human-to-human transfer has not been proven yet, S. pseudintermedius is currently on stage 2 according to Wolfe et al.
The abovementioned examples demonstrate that the list of zoonotic pathogens being monitored by the European Union epidemiological authorities should remain open, because evolutionary changes of many different pathogens occur incessantly. In the foreseeable future, particularly because potential animal reservoirs live close to humans, next animal pathogens might break the animal-human interspecies barrier and pose a substantial epidemiological risk. The importance of this problem is illustrated by the notable increase in the number of scientific publications on this subject.
Piśmiennictwo
1. Taylor LH, Latham SM, Woolhouse ME: Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 2001; 356: 983-989. 2. Woolhouse M, Gaunt E: Ecological origins of novel human pathogens. Crit Rev Microbiol 2007; 33: 231-242. 3. Wolfe ND, Dunavan CP, Diamond J: Origins of major human infectious diseases. Nature 2007; 447: 279-283. 4. Szewczyk EM: Kolejne patogeny zwierzęce poważnym zagrożeniem dla ludzi. Nowa Klin 2010; 17: 325-329. 5. European Food Safety Authority, European Centre for Disease Prevention and Control: The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2012. EFSA J 2014; 12: 20-175. 6. Koh TH, Sng LH, Yuen SM et al.: Streptococcal cellulitis following preparation of fresh raw seafood. Zoonoses Public Health 2009; 56: 206-208. 7. Weinstein MR, Litt M, Kertesz DA et al.: Invasive infections due to a fish pathogen, Streptococcus iniae. S. iniae Study Group. N Engl J Med 1997; 337: 589-594. 8. Baiano JC, Barnes AC: Towards control of Streptococcus iniae. Emerg Infect Dis 2009; 15: 1891-1896. 9. Sun JR, Yan JC, Yeh CY et al.: Invasive infection with Streptococcus iniae in Taiwan. J Med Microbiol 2007; 56: 1246-1249. 10. Zalas-Wiecek P, Michalska A, Grabczewska E et al.: Human meningitis caused by Streptococcus suis. J Med Microbiol 2013; 62: 483-485. 11. Lun ZR, Wang QP, Chen XG et al.: Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect Dis 2007; 7: 201-209. 12. Gottschalk M, Xu J, Lecours MP et al.: Streptococcus suis Infections in Humans: What is the prognosis for Western countries? (Part II). Clin Microbiol Newsl 2010; 32: 97-102. 13. Rantala S: Streptococcus dysgalactiae subsp. equisimilis bacteremia: an emerging infection. Eur J Clin Microbiol Infect Dis 2014; 33: 1303-1310. 14. Brandt CM, Spellerberg B: Human Infections Due to Streptococcus dysgalactiae subspecies equisimilis. Emerg Infect 2009; 49: 766-772. 15. Talan D, Staatz D, Staatz A et al.: Staphylococcus intermedius in canine gingiva and canine-inflicted human wound infections: laboratory characterization of a newly recognized zoonotic pathogen. J Clin Microbiol 1989; 27: 78-81. 16. Guardabassi L, Loeber M, Jacobson A: Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet Microbiol 2004; 98: 23-27. 17. Del Pace S, Savino A, Rasoini R et al.: A 72-year-old man with intermittent fever, anemia and a history of coronary and peripheral artery disease. Intern Emerg Med 2010; 5: 415-420. 18. Gerstadt K, Daly JS, Mitchell M et al.: Methicillin-Resistant Staphylococcus intermedius Pneumonia Following Coronary Artery Bypass Grafting. Clin Infect Dis. 1999; 29: 218-219. 19. Atalay B, Ergin F, Cekinmez M et al.: Brain abscess caused by Staphylococcus intermedius. Acta Neurochir 2005; 147: 347-348. 20. Durdik P, Fedor M, Jesenak M et al.: Staphylococcus intermedius – rare pathogen of acute meningitis. Int J Infect Dis 2010; 14: 236-238. 21. Cole LK, Kwochka KW, Kowalski JJ, Hillier A: Microbial flora and antimicrobial susceptibility patterns of isolated pathogens from the horizontal ear canal and middle ear in dogs with otitis media. J Am Vet Med Assoc 1998; 212: 534-538. 22. Kadlec K, Schwarz S, Perreten V et al.: Molecular analysis of methicillin-resistant Staphylococcus pseudintermedius of feline origin from different European countries and North America. J Antimicrob Chemother 2010; 65: 1826-1828. 23. Walther B, Hermes J, Cuny C et al.: Sharing more than friendship – nasal colonization with coagulase-positive staphylococci (CPS) and co-habitation aspects of dogs and their owners. PLoS One 2012; 7: e35197. 24. Stegmann R, Burnens A, Maranta C, Perreten V: Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. J Antimicrob Chemother 2010; 65: 2047-2048. 25. Riegel P, Jesel-Morel L, Laventie B et al.: Coagulase-positive Staphylococcus pseudintermedius from animals causing human endocarditis. Int J Med Microbiol 2011; 301: 237-239. 26. Laurens C, Marouzè N, Jean-Pierre H: Staphylococcus pseudintermedius and Pasteurella dagmatis associated in a case of community-acquired pneumonia. Mèdecine Mal Infect 2012; 42: 129-131.
