© Borgis - Nowa Medycyna 1/2009, s. 69-75
*Jürgen H. Helbig1, Junko Amemura-Maekawa2
Serotyping of Legionella pneumophila and epidemiological investigations
1Institute of Medical Microbiology and Hygiene, Medical Faculty of the Technical University Dresden, Dresden, Germany
2Department of Bacteriology, National Institute of Infectious Diseases, Tokyo, Japan
Streszczenie
Legionella pneumophila, the causative agent of Legionnaires´ disease, is characterised by a high degree of serological diversity caused on its lipopolysaccharide (LPS) structure.
Aim: Evaluation of L. pneumophila serotyping based on monoclonal antibodies (MAbs) of the Dresden Panel using ELISA and its possible employment in epidemiological investigations.
Material and methods: Patient and environmental isolates of L. pneumophila were typed using ELISA and/or indirect immunofluorescence test (IFT) according to standard procedures.
Results: The LPS epitope recognized by MAb 3/1 is virulence-associated. Its distributions among patient and environmental isolates differ significantly (P<0.01). Addition of MAb-typing to sequence typing of L. pneumophila increases the discrimination power.
Conclusions: Recognition of L. pneumophila LPS diversity using high-specific MAbs opens detailed information about taxonomy and Legionella populations in aquatic habitats. ELISA is the method of choice for serotyping of high numbers of isolates with many MAbs. Differentiation of Legionella strains into LPS profiles is an important step for confirmation or refutation of the identity of patient and environmental isolates and supplements additional information to genotypical data.

Introduction
Legionella pneumophila can be divided into at least 15 serogroups. Beyond it the so-called monoclonal subgroups exist within serogroups 1 (at least 10), 4 (two), 5 (two), and 6 (two). The serogrouping bases originally on the use of rabbit sera raised against the different serogroup reference strains which mostly be deposited at ATCC. The serogroup specificity of L. pneumophila is related to its lipopolysaccharide (LPS) characteristics (1). Since many of the serogroups have LPS antigens and house-keeping proteins in common, cross-reacting antibodies should be removed from the polyclonal immune sera by absorption with bacterial suspensions of all heterologous serogroups. Nevertheless, the success obtained depends on the individual immune sera and sometimes the reproducibility is not satisfactory particularly in comparative studies between different laboratories using different lots of antisera. Really, a sufficient kit for serotyping into the known serogroups is not available on the market.
About ten years ago it was obtained that each of the serogroups possesses a serogroup-specific epitope located on the LPS molecules detectable by using monoclonal antibodies (MAbs). These MAbs were summarized in the so-called Dresden Panel (2). Furthermore, MAbs recognising the monoclonal subgroups mentioned above were included in the panel (3). The established whole Panel is used in several European Legionella Reference Centres and other laboratories but also in Japan and the States.
Aims of the study
The first aim of this paper summarizes the own experience as well as the experiences of other laboratories regarding the use of the Dresden Panel for serotyping on the serogroup level and below it. In a second point the application of L. pneumophila serotyping by using MAbs for epidemiological investigations is discussed. Finally, the value of serotyping in the era of genotyping is supposed.
Material and methods
The hybridoma cell lines for producing MAbs were created in the Institute of Medical Microbiology and Hygiene, Dresden, Germany. MAbs were produced in cell culture flasks. For serotyping only cell culture supernatants containing approx. 20 ug/ml immunoglobulin were used. The working dilution depends on the MAbs and the assay used.
The strains were tested by ELISA and/or indirect immunofluorescence technique (IFT). The antigens for ELSA were prepared by scrapping the bacteria from BCYE agar plates, which were cultivated two or three days. The bacteria were heat-killed (10 min, 95°C), diluted in PBS to 1x108 bacteria per ml (approx. 0.3xMcFarland 1). The detailed procedure was described elsewhere (2). Shortly, coating of the wells overnight at 4°C, blocking of free binding sites of the wells with PBS containing 10% (v/v) fetal calf sera or 1% (w/v) BSA, addition of MAbs in working dilutions that have to be determined for each MAb. Bound MAbs were detected by anti-mouse Ig-horseradish peroxidase followed by substrate reaction with 3,3´,5,5´-tetramethylenbezidine/H2O2.
For IFT bacteria were diluted to McFarland 1 dropped on slides followed by air drying and heat-fixation (120°C). For evaluation of the serogrouping by MAbs strains were tested with home-made rabbit sera raised against the known serogroups followed by FITC-conjugated anti-rabbit mouse-Ig. For comparison of the MAb serotyping by ELISA and IFT, strains were also tested by MAbs. Bound MAbs were visualized by FITC-conjugated anti-mouse-Ig.
Results and discussion
Serotyping using MAbs
In a former study with 1064 L. pneumophila patient and environmental strains isolated in eleven European countries we have compared the serogrouping by ELISA based on serogroup-specific MAbs and the use of specific rabbit sera by IFT (2). Ninety eight percent of the strains could be placed into the known serogroups by using MAbs in most cases (97%), this was in agreement with the polyclonal typing by IFT. Of the29 isolates that showed strong cross-reactions with the rabbit antisera panel, 11 could be typed easily by MAbs, for the remaining 18, however, only serogroup-cross-reactive epitopes could be determined. Recently, altogether 50 of such atypical strains were further characterized by testing with five serogroup-cross-reactive MAbs resulting in additional nine phenons (4).
Below the serogroup level ten named monoclonal subgroups exist within the most important serogroup 1 (5). Out of these the Dresden Panel is able to differentiate eight subgroups on the same level (fig. 1). The other two subgroups were recognized as pool (3). It is to mention that below the subgroup level of L. pneumophila there exist additional serological variants.In general, the antigenic differences below the serogroup level can not be recognized by rabbit antisera.
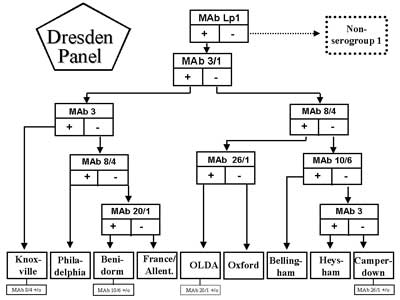
Fig. 1. Flowchart for monoclonal subgrouping of L. pneumophila serogroup 1 using the Dresden Panel. Serological variants among subgroups are added. +/o stands for positive/negative antigenic variants of monoclonal subgroups. Flowchart according to (3) was adapted.
Comparison of the usefulnessbetween the monoclonal serotyping by ELISA and IFT
MAbs were tested by both ELISA and IFT. Comparing the two methods, no significant differences in epitope recognition were seen. Good or bright fluorescence corresponds with an ELISA OD ≤ 0.5 and an OD of 0.2-0.5 is obtained in cases of weak fluorescence.
There are three main advantages of ELISA when compared with IFT. (i), decision about positive and negative reaction, respectively, bases on the cut-off level; (ii), high degree of reproducibility; (iii) carrying out of ELISA procedure can be done in parts mechanically which gives the possibility to serotype high numbers of isolates with many MAbs. Nevertheless, the use of the whole Dresden Panel for serotyping in one assay is not to recommend. In a first step it is well-tried to test with MAb Lp1 and the most common non- pneumophila serogroups 3 and 6. The second step splits into serogroup 1 subgrouping and the use of the other serogroup-specific MAbs supplemented with serogroup-cross-reactive MAbs for recognition of strains not belonging to the known serogroups. IFT is the technique of choice, if only a few isolates were typed.
Distribution of serogroupsand monoclonal subgroups among patient and environmental isolates
The diagnostic „Gold standard” for legionellosis is culturing of respiratory specimens on BCYE-agar plates, which allows subsequently the serotyping of the causal agent. However, only about 11% of cases in Europe were confirmed by culture (6). According to the guidelines of the European Working Group for Legionella Infections (EWGLI) cases of legionellosis were subdivided into community-acquired (CA), nosocomial-acquired (NA) and travel-associated (TA). With reference to all cases of the years 2000 to 2002, 37.9% were CA, 8.6% NA and 20.6% (TA). No subdivision was available for 32.9% (6). A pan-European study on culture-proven Legionnaires´ disease diagnosed in 14 countries has disclosed the dependence of the L. pneumophila serotype pattern from the category of infection (3). CA cases were caused predominantly by serogroup 1 strains (83.7%), the percentage for TA cases accounts to even 92.3%, whereas the value for NA cases accounts to only 64.1%. Most interesting, these differences increase if only special MAb-types of serogroup 1 were considered.
According to the flowchart for monoclonal subgrouping (fig. 1) the first step of subgrouping is the reaction with MAb 3/1. MAb 3/1-positive strains (the so-called Pontiac-like strains) can be typed by the Dresden Panel into four subgroups. MAb 3/1-positive strains cause 6.6 times more CA cases than MAb 3/1-negative cases, 13.4 more TA cases but only 2.6 times more NA cases. Figura(e) 2 presents the percentages of MAb3/1-positive and MAb 3/1-negative strains (non-serogroup 1 included) causing CA and NA legionellosis being acquired in different regions of Europe and Japan. Most interesting, in United Kingdom and Mediterranean 88% and 81% of CA plus NA Legionnaires´ disease were caused by MAb 3/1-positive strains whereas the percentage in Scandinavia amounts to only 51%. In Central Europe as well as Japan this percentage amounts to approx. 66% (fig. 2).
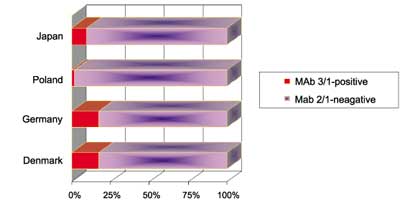
Fig. 2. Distribution of legionellosis caused by MAb 3/1-positive strains for different European regions [adapted according to (3) plus Japan (this study)].
The distribution of serotypes causing legionellosis differ from the data obtained for water systems, although representative data are available only for some countries (fig. 3). Nevertheless, the data are straight-forward. No more than 18% of the environmental isolates belong to the Pontiac-group. The individual percentages are significantly different (P<0.01%) from those of patient isolates in the corresponding countries Japan, Germany and Denmark.
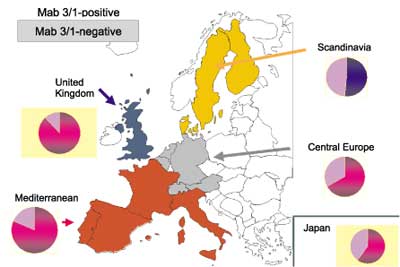
Fig. 3. Distribution of L. pneumophila MAb 3/1-positve strains in water systems from Japan (this study), Poland (Pancer, unpubl. data), Germany (7) and Denmark (8).
Taken together the epidemiological data mentioned above, the LPS structure that is recognized by the MAb 3/1 is virulence-associated. It is well accepted, that the extreme hydrophobicity of MAb 3/1-positive strains resulted in better survival conditions in aerosols. Nevertheless, it is not cleared up, if influence on host cell modulation exists, too.
Serotyping of L. pneumophilaas first step for confirmation of identity of patient and environmental isolates
The L. pneumophila populations in water systems are often not homogenous and more than one L. pneumophila strain is present. On the other side, sometimes different water systems can be assumed as the infective source. Therefore, it is important to recognize the whole population in order to find the causative agent of Legionnaires´ disease. That means, at least five agar-grown colonies of each water system should be serotyped. If there is among them no strain having the same serotype as the patient isolate, additional colonies have to be typed. The discrimination power of serotyping is low among MAb 3/1-positive patient isolates but relative high for all MAb 3/1-negative strains, which bring on, however, relative seldom a legionellosis. Nevertheless, Lück et al. (9) have described even a nosocomial pneumonia caused by three different strains of L. pneumophila belonging to two genotypes of serogroup 12 and to serogroup 1, MAb-subgroup Bellingham. All of them were isolated from the hospital water supply, too.
If on serological level the identity of patient and environmental isolates is confirmed, genomic investigations of the corresponding strains have to follow. Ten years ago the mostly used technique was pulsed-field electrophoreses. Since several years, sequence based typing (SBT) is the method of choice (see below). The strategy for confirmation of identity of patient and environmental isolates is summarized for TA cases in figure 4 [adapted from (10)].
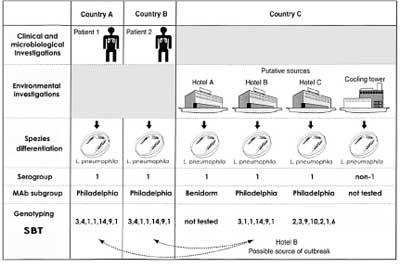
Fig. 4. EWGLI strategy for the epidemiological typing of strains examined on the investigation of travel-associated legionellosis [adapted to (10)].
Significance of serotypingon the side of sequence based typing (SBT)
The standard procedure of SBT recommended by EWGLI contains six gene targets comprising fliA, pilE, asd, mip, mompS, and proA.Ratzkow et al. (11) have been substantiated for 98 non-related L. pneumophila serogroup 1 strains that inclusion of neuA increases the discrimination index (DI). If MAb typing is included additionally, the DI is higher for both numbers of sequenced gene targets (tab. 1).
Table 1. Discrimination values (DI) and Konfidenzintervalls (KI; 95%) for standard-SBT (six genes) and standard plus neuA depend on MAb-typing for 98 non-related L. pneumophila serogroup 1 strains [summarized data from (11)].
| SBT | Inclusion of MAb-typing | DI-value | KI |
| Standard | no | 0.932 | 0.913-0.951 |
| Standard | yes | 0.974 | 0.968-0.980 |
| Standard plus neuA | no | 0.963 | 0.952-0.974 |
| Standard plus neuA | yes | 0.980 | 0.974-0.985 |
Powyżej zamieściliśmy fragment artykułu, do którego możesz uzyskać pełny dostęp.
Mam kod dostępu
- Aby uzyskać płatny dostęp do pełnej treści powyższego artykułu albo wszystkich artykułów (w zależności od wybranej opcji), należy wprowadzić kod.
- Wprowadzając kod, akceptują Państwo treść Regulaminu oraz potwierdzają zapoznanie się z nim.
- Aby kupić kod proszę skorzystać z jednej z poniższych opcji.
Opcja #1
29 zł
Wybieram
- dostęp do tego artykułu
- dostęp na 7 dni
uzyskany kod musi być wprowadzony na stronie artykułu, do którego został wykupiony
Opcja #2
69 zł
Wybieram
- dostęp do tego i pozostałych ponad 7000 artykułów
- dostęp na 30 dni
- najpopularniejsza opcja
Opcja #3
129 zł
Wybieram
- dostęp do tego i pozostałych ponad 7000 artykułów
- dostęp na 90 dni
- oszczędzasz 78 zł
Piśmiennictwo
1. Ciesielki CA, Blaser MJ, Wang WLL: Serogroup specificity of Legionella pneumophila is related to lipopolysaccharide characteristics Infect Immun. 1986; 51: 397-404. 2. Helbig JH et al.: The antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups J Clin Microbiol. 1997; 35: 2841-2845. 3. Helbig JH et al.: Pan-European study on Legionnaires´ disease: Distribution of Legionella pneumophila serogroups and monoclonal subgroups Eur Clin Microbiol. 2002; 21: 710-716. 4. Helbig JH et al.: Identification and serotyping of atypical Legionella pneumophila strains isolated from human and environmental sources J Appl Microbiol. 2002; 102: 100-105. 5. Joly JR et al.: Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies J Clin Microbiol. 1986; 23: 768-771. 6. Joseph CA: Epidemiology of Legionnaires´ disease in Europe 2000-2002 Epidemiol Infect. 2004; 132: 417-424. 7. LŘck PC, Helbig JH: Typing of Legionella strains isolated from patients and environmental sources in Germany, 1990-2000. In: Legionella. (Ed. Marre R et al.) ASM Press, Washington, DC, 2002 pp. 267-270. 8. Pringler NP, Brydov P, Uldum S A: Occurrence of Legionella in Danish hot water systems. In: Legionella. (Ed. Marre R et al.) ASM Press, Washington, DC, 2002 pp. 298-301. 9. LŘck PC, Wenchel H-M, Helbig JH: Nosocomial pneumonia caused by three genetically different strains of Legionella pneumophila and detection of these strains in the hospital water supply J Clin Microbiol. 1998; 36: 1160-1163. 10. Fry NK et al.: A multicenter evaluation of genotypic methods for the epidemiologic typing of Legionella pneumophila serogroup 1: results of a pan-European study Clin Microbiol Infect.1999; 5: 462-477. 11. Ratzow S et al.: Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 trains J Clin Microbiol. 2007; 45: 1965-1968. 12. Borchardt J, Helbig JH, LŘck PC: Occurrence and distribution of sequence types among Legionella pneumophila strains isolated from patients in Germany: common features and differences to other regions of the world Eur J Clin Microbiol Infect Dis. 2007; 27: 29-36. 13. ThŘrmer A et al.:PCR-based ´serotyping´ of Legionella pneumophila J Med Microbiol. 2009; 58: (in press).





